Chapter 9 Exercises
9.1 Introduction
These exercises are meant to provide practice of the course material presented in this eBook. There is no single good solution for them, but some are better than others. A good practice is also to try to solve the same exercise in several different ways.
The exercises have been marked with a difficulty level, from 1 to five “stars” (❋ to ❋❋❋❋❋). Since I am a big fan of intellectual challenge I have included quite some exercises that transcend the mastery level required at the final assessment. For your peace of mind; if you are able to solve three-star exercises you are really good to go!
Most of the datasets referred to from within the exercises can be found in the Github repository with the URL https://github.com/MichielNoback/datasets. This is a direct link. You can download individual datasets from this page, or download the entire repository at once. To download, use the “clone or download” pull down button (green button). If you want to be a pro, use git to clone it…
The solutions to the exercises are in the next chapter of this eBook.
For all plotting exercises: take care of the details: axis labels, figure title or caption clear names of separating variables.
9.2 The ggplot2 and tidyr packages
9.2.1 Trees
The datasets package that is shipped with R has a dataset called trees.
A [❋]
Create a scatter plot for Heigth as a function of Girth, with blue plotting symbols .
B [❋❋]
Add a single line representing the linear model of this relationship, without shaded confidence interval.
C [❋❋]
Have the Volume variable reflected in the size of the plot symbol and change their transparency level to 0.6.
9.2.2 Insect Sprays
The datasets package that is shipped with R has a dataset called ?. Type ?InsectSprays to get information on it.
A [❋]
Create a boxplot, splitting on the spray type.
B [❋❋]
Create a jitter plot, splitting on the spray type. Have zero jittering on the y-axis and as little as possible on the x-axis. use the alpha parameter and try to find a nice plot symbol.
[Extra: Give each spray a different plot color]
9.2.3 Diauxic growth
In 1941, Jacques Monod discovered the phenomenon of diauxic growth in bacteria (e.g. E. coli), resulting from the preference for some carbon substrate over others, causing catabolite repression of pathways of less preferred substrates. See The Wikipedia page for details.
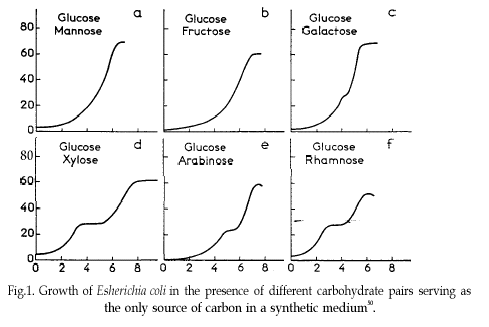
Diauxic growth (figure from wikipedia)
Some of the data used to generate the figure in that original publication are in this course’s data repository (https://github.com/MichielNoback/datasets/diauxic_growth). A direct link to the data file is https://raw.githubusercontent.com/MichielNoback/datasets/master/diauxic_growth/monod_diauxic_growth.csv.
A [❋❋]
Download the file, load it and attach to variable diauxic. Next, tidy this data into long format so you have three columns left: Time, Substrate and OD.
B [❋]
Convert the newly created Substrate variable into a factor with nicer labels, i.e. better suited for human reading
C [❋❋]
Create a line plot with all four growth curves within a single panel. use stat_smooth() and its span = parameter to have a line-and-point visualization.
D [❋❋❋]
Create a multi-panel plot like the one from the original publication.
9.2.4 Virginia Death Rates
The datasets package that is shipped with R has a dataset (a matrix) called VADeaths. Type ?VADeaths to get information on it. First you should convert this matrix into a tibble (a kind of data frame) and the rownames into a real variable using the chunk below (study it so you understand what happens here):
library(dplyr)
## %>% is used to pipe results from one operation to the other, just like '|' in Linux.
virginia_death_rates <- as_tibble(VADeaths)
virginia_death_rates <- virginia_death_rates %>%
mutate("Age Group" = factor(rownames(virginia_death_rates), ordered = TRUE)) %>%
select(`Age Group`, everything()) #reorder the columnsA [❋❋❋]
Pivot this table to long (tidy) format. This should generate a dataframe with four columns: Age Group, Habitat, Gender and DeathRate.
B [❋❋]
Create a bar chart with the age groups on the x-axis and Rural/Urban and Male/Female using side-by-side bars.
9.2.5 Investigate new visualization [❋❋❋]
Go to the R graph Gallery and browse the different sections. Then select one visualization to study in-depth (excluding the ones already demonstrated in this eBook). Work out an example with one of the datasets from either the built-in datasets of R or the Datasets Repo of this course. You can also choose another dataset. For instance, Kaggle (https://www.kaggle.com/) and the UCI Machine Learning Repository (https://archive.ics.uci.edu/ml/) have very interesting datasets.
Present this visualization in class, addressing these topics:
- What is this visualization named?
- What is its purpose; when is it appropriate to use?
- Why does it appeal to you?
- Show an example with the R code, explain difficulties and point out caveats.
9.2.6 ToothGrowth
[❋❋]
The datasets package that is shipped with R has a dataset called ToothGrowth. Create a visualization distinguishing the tooth growth between both supplement and dose.
9.2.7 Puromycin
[❋❋❋]
Create a scatter plot visualization of the Puromycin dataset. Color by state and add a loess model smoother without error margin. Use the formula = parameter in geom_smooth() for getting a better-fitting regression line.
9.2.8 Global temperature
Maybe you have seen this picture of the world temperature over the past 120 years in the media:
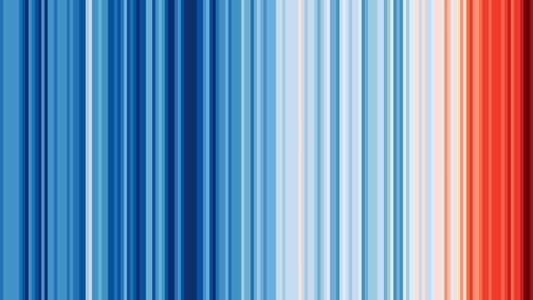
global_heatmap_s.png
We are going to work with this data as well.
The global temperature data are located in folder global_temperature (see the Data Repo).
There are two data series in file https://raw.githubusercontent.com/MichielNoback/datasets/master/global_temperature/annual.csv.
Study the readme file in the same folder to find out more about these datasets.
9.2.8.1 Create a scatter-and-line-plot [❋❋]
Create a scatter-and-line graph of both series in a single plot. Annotate well with labels and title. Optionally, add a smoother without error boundaries.
9.2.8.2 Re-create the heatmap [❋❋❋❋]
You should try to reproduce the above picture by using the geom_tile() function. Hint: use scale_fill_gradient2(low = "blue", mid = "white", high = "red") and pass 1 as value for y in the mapping function. The theme() function with element_blank() can be used for extra tweaking.
9.2.8.3 Extra practice [❋❋]
As extra practice, you could try to answer these questions as well:
- what is the warmest year?
- what is the warmest year that both timelines agree upon?
- which was the coldest decade?
- what is the 30-year moving average of the temperature?
The is also a monthly temperature file. Many explorations can be carried out on that one as well.
9.2.9 Epilepsy drug trial
The epilepsy folder (see the Data Repo) contains two files, one of which -epilepsy.csv- is the actual data file. The readme.md file describes the dataset and the columns. Read it carefully before proceeding.
9.2.9.1 Load the data [❋]
Load the data and be sure to check the correctness of data types in the columns. The period should be a factor. You can use a downloaded copy, but a direct link to the data file can also be used as argument to read.table():
https://raw.githubusercontent.com/MichielNoback/datasets/master/epilepsy/epilepsy.csv
9.2.9.2 Reorganize the data [❋❋]
Reorganize the data so that the dependent variable comes last and the useless entry variable is omitted. This order of variables is required:
"subject", "age", "base", "treatment", "period", "seizure.rate"
For better readability you should convert the dataframe into a tibble using as_tibble() (tibbles are dealt with in a later chapter: dplyr).
9.2.9.3 Create plots of seizure rates
A [❋❋]
First, create a boxplot of the seizure rates of both groups, split over both the period and the treatment. To support both of these data dimensions you will either have to use the color aesthetic, or the facet_wrap() function, like this + facet_wrap(. ~ treatment). I suggest you try them both.
B [❋❋❋]
Next, create the same basic plot as a jitter plot. You can use the facet_wrap(). Compare them and write down some pros and cons of both.
C [❋❋❋]
Investigate whether an overlay may improve the story this visualization tells, or can you come up with an even better graph?
9.2.9.4 A boxplot after correction [❋❋❋]
The base column is the base seizure rate of the subject in an 8-week window prior to the actual trial. To compare before and after you should create a new dataframe where the seizure.rate is summed for the 4 periods. Next, create a final boxplot of these corrected values.
9.2.9.5 Test for statistical significance [❋❋❋]
In the previous exercise you have determined the corrected seizure rates. Can you figure out a statistical test to see if the difference is significant?
9.2.9.6 Investigate age-dependency [❋❋❋]
Investigate whether there is an age-dependent effect in either the base seizure rate or the effect of the treatment.
9.2.10 Bacterial growth curves with Varioscan
Chapter 10, @ref(parsing-complex-data ), ended with a file being saved, data/varioscan/2020-01-30_wide.csv. This file is also present in the varioscan folder of the datasets repo.
9.2.10.1 Load, preprocess and tidy [❋❋❋❋]
First load the data; this is straightforward. Next, correct OD values of the “triplicate” experiments using the background signal data from samples without bacteria.
Thus, the Red values should be corrected using the Red_w_o values of the same dilution; the White columns using White_w_o and Elution using Eluiton_w_o.
This can be done when you apply a trick: transpose the dataframe using t() before carrying out the corrections. After transpose, you should loop the columns and correct using the periodicity of 8 (number of dilutions)
You can use this as a start, assuming you loaded the file into growth_data:
## Add rownames to get a hold of them after transpose
rownames(growth_data) <- paste0(growth_data$Content, '.',
growth_data$.copy, '.',
rep(LETTERS[1:8], times = 12))
## transpose
tmp <- t(growth_data)
## build a new dataframe/tibble
growth_data_corr <- tibble(column_names = rownames(tmp))
## continue hereAfter these steps, the data should be tidied.
9.2.10.2 Create a growth curve visualization [❋❋❋❋]
Create a line plot visualizing the entire experiment. The challenge here is to present a clear picture with tons of dataseries on top of each other. You should realise there are 96 time series that could be plotted.
I suggest you also use a custom color pallette reflecting the sample groups in the experiment.
The main sample set (the red tulip) should be emphasized. The least interesting controls, columns 4, 8 and 12, should be made less present visually.
9.2.11 The dinos
This exercise represents a review of many of basic R operations as well, besides ggplot2.
The dinos folder contains an Excel file called jgs2018049_si_001.xlsx. It contains supplementary data to a scientific publication with the title “The Carnian Pluvial Episode and the origin of dinosaurs” [Benton et al., Journal of the Geological Society 175(6) 2018].
One of the figures accompanying the paper is this one:
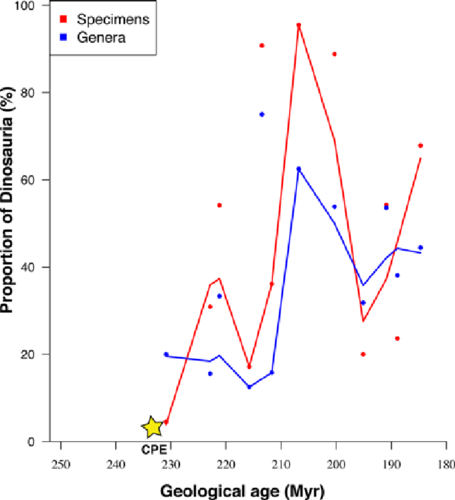
Figure 2.Proportions of early dinosaurs through the Triassic, showing the rapid rise in the late Carnian and early to middle Norian. Two metrics are shown, numbers of specimens and numbers of genera in 12 sampled faunas, in proportion to all tetrapods; the linking line is a moving average. Specimen counts perhaps exaggerate the trend when compared with generic counts, or at least both show different aspects of the same rise in ecological impact of the dinosaurs in the Late Triassic. (Based on data in supplementary material Table S1.)
You have to agree this is terrible! They even omitted the legend for CPE which is descried in the paper “Carnian Pluvial Episode (CPE), dated at 232 M Ya.” And what the heck is a “moving average?”
Let’s explore and improve.
9.2.11.1 Export to csv
We’ll begin by exporting all three tabs to a textual format.
Open the excel file and select the tab “Contents.” You can see it is a “codebook” - it contains column names and descriptions for the two other tabs in the excel file. In the File menu, select Save As…. Next, give as name codebook.csv and for File Format, select CSV UTF-8 (Comma-delimited) (.csv). You’ll get a warning -read it!- but select OK.
Next, select tab “Skeletons” and repeat to generate skeletons.csv and “Footprints” to generate footprints.csv.
Close the Excel file without saving changes to it. We’ll continue with the csv files.
Note: R has packages providing functionality to read from Excel, but this is outside the scope of this course, and installing them is often a hassle.
9.2.11.2 Clean up and load codebook.csv
Have a look at the contents of codebook.csv in the editor. It is not yet suitable for loading into R. Write down the number of lines describing column headers for the skeletons and the number for footprints. Next, delete the lines that are not really codebook entries.
A [❋]
Load the resulting file -as character data only!- and assign it to variable codebook.
B [❋❋]
Give the dataframe column names: variable and description.
C [❋❋]
Remove the leading space of the second column
D [❋❋]
Add a column called dataset: a factor with the value skeleton or footprint, depending on the file that is referred to.
9.2.11.3 Write a utility function [❋❋❋]
Write a utility function that returns a text label to be used in plotting when given a dataset name and a column name. The label should come from the codebook description variable of course. The dataset parameter should default to skeleton. For example, these calls:
get_description('Dinosaur_gen', 'skeleton')
##same as
get_description('Dinosaur_gen') should both return Number of genera of Dinosauria.
As extra challenge you could implement some error checking to make the function more robust.
9.2.11.4 Load skeleton data [❋]
Load the data in the skeletons.csv file and assign it to variable skeleton. Make sure your data columns have the right type and that you did not overlook NA value or decimal encodings.
9.2.11.5 Plot species versus time [❋❋❋]
As a first exploration of the data, create a scatterplot of Total_spec as a function of Midpoint (the midpoint of the archaeological epoch) and have the points colored by Epoch. You should use ggplot2 of course.
There is an outlier flattening the picture quite a lot. Can you think of a strategy to make the picture clearer?
Another problem is that the x-axis scale is from recent to ancient and this should be reversed.
Finally, add a smoother (loess regression) for the entire dataset (not split over the Epochs!) and annotate the plot with nice axis labels, preferably using your previously created utility function. NB: it may be a good idea to tweak the descriptions in the codebook a little bit.
Note that for “Million Years Ago” you can use the abbreviations “MYA” or “Ma” (Mega annum).
9.2.11.6 Reproducing the publication figure
Reproduce the figure from the introduction of this section, but using ggplot2 instead of base R as they used. This requires some preprocessing steps, especially with the use of pivot_longer().
A [❋❋]
You will have to calculate the proportions of Dinosauria (Dinosaur_gen) relative to the total of all “tetrapod” groups (including Archosauromorph_gen, Dinosaur_gen, Synapsid_gen, Parareptile_gen, Temnospondyl_gen). You need this for Specimens (_spec) as well as Genera (_gen).
B [❋❋❋]
Next, you should extract a single proportion for all Formations represented in a single Midpoint. I realize that this is not an entirely valid operation. Do you know why? Can you figure out how it was done for the existing publication plot?
I suggest you use the aggregate() function. You will need the Epoch and Stage of these as well for later aspects of plotting.
C [❋❋❋]
Now you need to tidy the data.
D [❋❋❋❋]
Finally you have the data to generate the plot itself, without the moving average.
E [❋❋❋❋❋]
Add the moving average. A simple moving average (SMA) is the unweighted mean of the previous n data.
The paper does not say anything about the “window” (n) used in the moving average, but looking at the original figure it is less that 10 MY wide. Actually, to be honest, I cannot identify the used algorithm by looking at the plot. Can you?
Just give it a shot and see how far you get.
9.2.11.7 Make a better figure [❋❋❋]
Next, try to make a plot that uses an alternative to the moving average used in the publication. Also, use background coloring (rectangles) to highlight the Epochs within the plot.
9.2.11.8 Use the size aesthetic [❋❋❋❋❋]
The “size” aesthetic can be used for indicating the absolute number of specimens/genera, respectively. This adds an extra dimension of information to the plot.
9.3 The tidyr and dplyr packages
9.3.1 Global temperature revisited
Reload the global_temp data if it is not in memory anymore.
A [❋❋]
Which years before 1970 had an anomaly of more than +0.1 degree according to the GCAG model? And according to the GISTEMP model? Notice anything strange?
B [❋❋]
Which has (have) been the coldest year(s) after 1945? Do both models agree on that?
C [❋❋❋]
Which were the warmest five years? Select the warmest five for both models. Do they agree on that?
To select the top n values after grouping, use filter(row_number() %in% 1:5)
D [❋❋❋]
Select the years from 1970 and split up the dataset in decades and report the per-decade average temperature for both models.
Extra exercise: create a bar plot from this result.
9.3.2 ChickWeight
You have seen how to determine the weight gain of the chickens between measurements in the ChickWeight dataset. Here, we look at this dataset again.
Starting weight [❋❋].
Which chick had the highest starting weight? List this chick, together with its diet as follows:. Total weight gain [❋❋❋].
Determine the total weight gain for each chick and list all chickens together with their diet and number of weight measurements. Order from high to low weight gain.Average weight gain per diet [❋❋❋❋]. Determine the average weight gain for each diet. Report the Diet, number of chickens on the diet and the average weight gain.
9.3.3 Population numbers
The population folder of the datasets repo contains two files. One of them, EDU_DEM.....csv contains population data for all countries of the world (where data is available). We’re going to mangle and explore this data a bit.
Besides the following exercises there are many, many other questions you could ask yourself about this dataset, and visualizations to be created. For instance, how about countries and age groups, years with maximum growth, etc. Explore and practice!
9.3.3.1 Load the data [❋]
Start by loading the data into a dataframe called population.
Since it is a bit large, I suggest you first download it to a local location.
9.3.3.2 Clean up [❋❋]
There are quite some uninformative variables - variables with only one value or with only NA. Find out which variables are uninformative and remove these from the dataframe.
9.3.3.3 Create a “wide” yearly report of population totals [❋❋❋]
With the dataframe you now have, create a report in wide format for the total population numbers over the available years. So, the years now get their own column and each country has a single row.
9.3.3.4 Create a “wide” yearly report of population change [❋❋❋❋]
Next, present the same format (wide), but now with the “population change” instead of the total population.
9.3.3.5 Create a bar plot split on gender [❋❋❋]
Create a bar plot of the population numbers of the countries of Western Europe across the available years, split by gender.
9.3.3.6 Highest growth rate [❋❋❋]
Which three countries have shown the fastest relative growth rate, measured as percentage change over the the whole of the available time period (2005-2017) relative to its start, over the entire population, for both sexes combined?
Extra practice: Do the same for the different age groups.
9.3.4 Rubella and Measles
The rubella_measles_cases folder of the datasets repo contains historic data for both measles and rubella. This serves as extra training material (no solutions provided). Try to report, both visually and in tables:
- cases per per continent
- cases over time per continent
You can also combine with the population data from a previous exercise (folder population) and report measles and rubella per 10000 inhabitants, per country or per continent.
Other data are available for exploring relations as well in the population data: age groups, sex, and diseases.
9.4 The lubridate package
These exercises can of course only be completed if you also apply your knowledge of the tidyr and dplyr packages.
9.4.1 Female births
The datasets repo contains a folder female_births. Its content is fairly simple: it holds the number of daily female births in California in 1959. We’ll have a look at it now.
9.4.1.1 Load the data [❋❋❋❋]
Use local caching for this dataset using conditional exectution of download.file(). Load the data and use the lubridate package so that the Date column becomes a Date type.
9.4.1.2 Check for missing rows [❋❋❋]
Check if there is really not a single day missing in this reported year. Hint: one option is to look at the lag() between subsequent rows.
9.4.1.3 Report birth numbers [❋❋❋]
Report the number of births per month (Jan - Dec) and per weekday (Mon - Sun), as barplots. Is there a month or day that seems to be anomalous, or do you see seasonal trends? Try a statistical test!
9.4.2 Diabetes
The datasets repo of this course contains a folder named UCI_diabetes_dataset. The complete dataset contains various types of information for 70 diabetes patients. Study the Readme file of this dataset before proceeding.
9.4.2.1 Create a codebook [❋❋]
Copy-and-paste the Code Fields into a file named codebook.txt and load it as a dataframe with name codebook_diab.